Does Flow Play A Role in Protein Aggregation During Bioprocessing?
By Leon F. Willis, Sheena E. Radford, Nikil Kapur, and David J. Brockwell, University of Leeds

Most bioprocessing steps involve fluid flows, from mixing during fermentation through to the fill/finish of the drug product. The aggregation and degradation of therapeutic proteins during processing has long been of concern to the field,1 particularly as aggregates can trigger adverse immunogenic effects in vivo,2,3 decreasing product yield and increasing cost of goods.4 While the mechanisms of, e.g., pH5 and temperature-induced protein aggregation6 are well-known, it is less clear how and indeed, if, fluid flows induce protein aggregation. In this article, we wish to highlight recent developments in the field that address this.
Hydrodynamic Forces And Aggregation: More Than Just ’Shear’
To dissect the potential mechanisms underpinning flow-induced protein aggregation, the hydrodynamic environment encountered by a protein needs to be broken down into its constituent parts. Many in the bioprocessing field refer to the potentially harmful effects of fluid flow on proteins as “shear forces”.7,8 “Shear” has a specific definition in fluid mechanics, namely the superposition of rotational flow and elongational/extensional flow. Specific flow types exist within certain hydrodynamic environments, e.g., shear flow dominates within laminar flows within a pipe (Figure 1a), whereas extensional flows may play a role within other parts of the process - for example, fill/finish. Extensional flows occur when fluids are forced to accelerate in the direction of the flow, often caused by a constriction in the flow path, e.g., passing through a filter pore (Figure 1b).
We define protein aggregation as the self-association of “non-native” (i.e., partially or fully unfolded) protein species.9,10 It has long been thought that shear flow preserves the native structure of globular proteins irrespective of the magnitude of the shear rate and the total exposure time, as proteins essentially “roll around” as they tumble in shear flow in free solution.11 Unless the protein’s topology is elongated and/or has a molecular weight >150 kDa, where drag forces (most significant near the walls) can cause conformational rearrangement,12,13 the shear rates necessary to perturb secondary structural elements are thought to be high (exceeding 104 s-1).14–16 Consequently, shear flow has been discounted as being capable of inducing protein aggregation.15,17 Extensional flow, on the other hand, imparts strain on molecules. It has been shown in simulations for the topologically elongated blood protein von Willebrand factor that elongation of the protein molecule is observed at strain rates an order of magnitude lower than shear rates.18,19 Depending on the protein (some of which are evolved to respond to fluid flow),20,21 either in a pure extensional flow or the extensional component of a shear flow, strain may stretch out the protein structure, exposing previously buried aggregation-prone regions to solution, with protein aggregation then proceeding if these species self-associate under the conditions employed (Figure 1c).22
Finally, both solid-liquid and air-liquid interfaces are presented to proteins during processing (Figure 1d), with these being increasingly linked to protein aggregation under flow.23,24 Proteins can readily adsorb to these interfaces, potentially leading to their denaturation or misfolding and aggregation.25 The order in which these steps occur, together with the role fluid flow plays in interfacial renewal, is still the subject of debate in the field.24 Furthermore, at solid interfaces, shear flow imparts force at the wall, defined as shear stress (Figure 1d).26 In vivo, these forces trigger mechanobiological events.27 In vitro, it has been shown that shear stress can also bring about conformational changes in proteins.28
As both bulk fluid flow and interfacial stresses are important in the mechanisms governing protein aggregation, we suggest that “hydrodynamic forces” serves as a more appropriate umbrella term than “shear” when discussing processing-induced protein conformational change and aggregation.
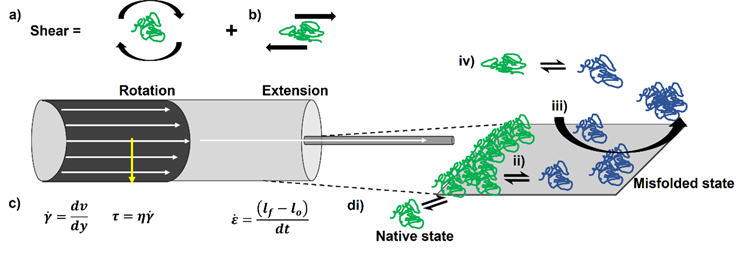
Figure 1. Properly defining hydrodynamic forces identifies potential aggregation mechanisms for proteins under flow. a) Considering flow in a pipe, the fluid moves fastest at the center and slowest at the wall, due to friction. The difference in velocity perpendicular to the direction of flow (dv (m.s-1) divided by the difference in distance between the lamellae (layers) of fluid (dy, m) allows the shear rate (y,s-1) to be defined. Given the dynamic viscosity of the fluid (η, Pa.s), the shear stress τ (Pa) can be calculated. b) Where constrictions occur in a flow, the fluid is forced to accelerate to overcome the constriction, generating an extensional flow. A fluid element or molecule may elongate in this flow field, thus its final length lf will be greater than its original (lo) defining strain, ε. Extensional flows are quantified from the change in strain over time (dt) = strain rate (s-1). c) Pure shear flows consist of rotational and extensional elements. d) No fluid flow field exists in the absence of an interface. For the solid-liquid interface in the pipe, natively folded proteins (green) could adsorb to the surface (i). This may cause conformational changes in the protein, which expose their previously buried hydrophobic regions(ii). The misfolded species (blue) could then aggregate, with flow removing them from the surface (iii). Bulk flow may also trigger these conformational changes (iv) but its importance to manufacture-induced aggregation has been questioned.
Where Are Hydrodynamic Forces Found In Processing?
Hydrodynamic forces are encountered by therapeutic proteins throughout their lifetime. In upstream processing (Figure 2a), stirring from impellers generates shear flows,29,30 with air-water interfaces in the form of bubbles and cavities (vapor-filled gaps in the fluid) also forming during fermentation. These could impact the protein product through the mechanisms described above, as well as potentially damaging the mammalian cells used to over-express proteins of interest.31
During downstream processing (Figure 2b), different solid surfaces are encountered during chromatography (e.g., different resin chemistries32 and pH changes5), depth,33 and ultrafiltration.34 The move to single-use technologies has meant that an array of polymers (e.g., Puraflex bags, PTFE, and elastomers) are employed.35 The scale of these operations (in terms of surface area to volume ratio and volumetric flow rate) will of course vary depending on the amount of quantity of material being processed. The duration of these unit operations will affect the relative exposure time of proteins to the different hydrodynamic forces involved in each step and, given that aggregation is a time-dependent process, can be critical in determining protein behavior in different devices.
Fill/finish, where the drug substance goes into, e.g., a vial or prefilled syringe, often occurs at fast flow rates (several mL/s), exposing protein solutions (often at high concentrations) to high shear rates (exceeding 105 s-1)15,36,37 and extensional flows (Figure 2ci). As these final steps turn drug substance into the (most valuable) drug product, aggregation at this critical stage would adversely affect the cost of goods.
Finally, the drug product is transported to the patient, with shaking during transport potentially generating shear flows and air-water interface turnover, which could drive aggregation in the vial.38,39 Subjecting protein solutions to a mechanical shock, e.g., dropping a vial or syringe, has been shown to cause cavitation, which could adversely impact the product prior to delivery into the patient (Figure 2cii and ciii).38
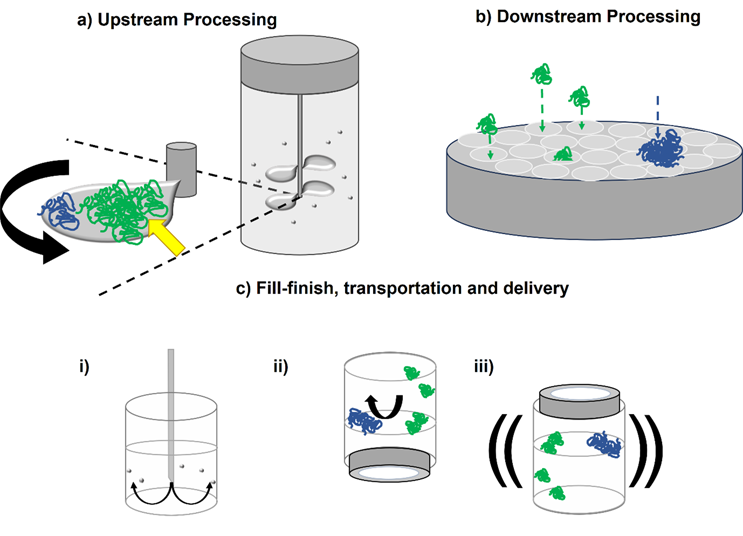
Figure 2. Hydrodynamic forces in particular unit operations may induce protein aggregation. a) In upstream processing, e.g., fermentation, proteins could adsorb to the rotating impellers and/or cavities. As well as these surfaces facilitating conformational change to the protein (green to blue), high shear rates at the impeller tip and shear stress on the surface (yellow arrow) may drive misfolding and aggregation. b) In downstream processing, e.g., dead-end filtration, extensional flows will be present as proteins navigate the membrane’s pores. Protein aggregates may block the pores, with fouling increasing shear stress on the membrane. ci) In fill/finish, vials can be filled with high concentration protein drug substance through narrow gauge needles. The resulting extensional flow and potential cavitation of the fluid could drive aggregation. cii) Air-water interfaces found in the headspace could drive aggregation in a vial, with shaking (iii) exacerbating their turnover.40,41 Surfactants are commonly added to biopharmaceuticals to prevent these harmful adsorption events taking place.
How Are The Effects Of Hydrodynamic Forces On Proteins Studied?
Over the past 40 years researchers have developed an array of tools to interrogate whether hydrodynamic forces induce protein aggregation. These are summarized in Figure 3.
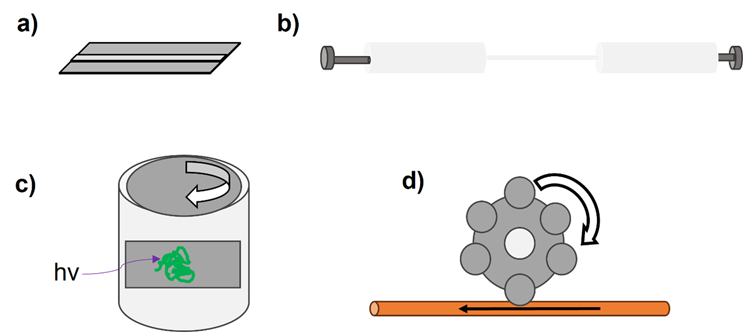
Figure 3. Schematic representations of different devices used to exert hydrodynamic forces on proteins. a) Microfluidic devices have been used to evaluate shear stresses on proteins such as serum albumin and antibodies.28,42 b) Following our development of an extensional flow device (EFD) in 2017,16 many groups have since built similar syringe-based systems to exert varied hydrodynamic forces on proteins.43–46 c) Couette rheometers have been adapted by many to exert shear flows on proteins,47 as their configuration lends itself to the integration of different analytics, e.g., circular dichroism (CD) spectroscopy.48 d) Peristaltic pumps have been used to evaluate the influence of tubing material on protein aggregation.49
Some researchers have repurposed scientific equipment, such as rheometers, to probe the effects of shear flow on proteins.15 Others have built bespoke devices to probe the effects of shear flow,50,51 shear stress,17,28,52 extensional flow, and interfaces.16,41,43–46 Of these, some instruments were designed to explicitly mimic bioprocessing steps, e.g., pumping at small (
The diversity of proteins and the infinite possibility of solution and formulation conditions that have been used, together with the wide array and depth of characterization of the devices used to subject proteins to hydrodynamic forces (e.g., extensional flow was historically neglected by the field), have all contributed to the lack of complete understanding of the fundamental problem of how flow stress affects proteins and how best to avoid this in bioprocessing. While an overemphasis on “shear” may have contributed to this issue, the field seems to have reached a consensus that a combination of both flow and interfaces are important with respect to how hydrodynamic forces induce protein aggregation, with the underlying mechanism often being protein specific.7,24,43
Can We Control Aggregation Under Flow?
In this article, we have defined what hydrodynamic forces are, discussed the mechanisms by which they could cause protein aggregation, and highlighted where flow-induced problems could occur in bioprocessing. Having surveyed the practical tools available to investigate these phenomena at small scale, one may wonder what can be done to protect proteins from these potentially damaging effects?
Firstly, interfaces are encountered throughout bioprocessing and are both ubiquitous and varied in nature. Certain materials may be protective or damaging to proteins, depending on the nature of the protein-interface interaction (Figure 1b). Materials can be changed in some circumstances, with single-use technologies offering more options than often-used stainless steel. The choice of tubing material has been shown recently to be particularly important for some applications.58 Biopharmaceutical formulations routinely include surfactants to protect against interfacial stresses.59 However, there are many caveats to the choice and mechanism of action of these additives, which are outside the scope of this article (see60,61 ). Other excipients that protect against bulk aggregation include co-solutes such as sucrose62,63 or protective amino acids such as arginine.64,65 Literature reports, including from our own group, show that arginine can suppress antibody aggregation under flow.56 The development of small-scale flow devices has enabled industrial researchers to evaluate how these formulation additives impact their products. We hope that our research and this report will help industry protect the valuable protein drugs of the future against the potentially harmful effects of flow-induced stress, buoyed by the recent surge in research activity in this field by scientists across the world.
The authors wish to thank the following for funding and supporting our research in this area over the past decade: The Royal Society (RSRP\R1\211057), Wellcome Trust (WT204963), European Research Council (FP7/2007-2013 Grant Agreement 32240), EPSRC (EP/I033270/1 and EPSRC Doctoral Prize Fellowship to LFW), The Royal Academy of Engineering, our industrial collaborators past and present (AstraZeneca PLC, GlaxoSmithKline PLC, Pfizer LLC, Adimab LLC, Malvern Panalytical PLC), and the University of Leeds. Disclaimer: none of the funders were involved in the writing of this article.
About The Authors:
 Leon Willis, Ph.D., is a postdoctoral researcher at the University of Leeds’ Astbury Centre for Structural Molecular Biology in the School of Molecular and Cellular Biology. Since 2014, his work has focused on how hydrodynamic forces can damage proteins, specifically, monoclonal antibodies. He studied biological chemistry at the University of Sheffield and obtained his Ph.D. at Leeds.
Leon Willis, Ph.D., is a postdoctoral researcher at the University of Leeds’ Astbury Centre for Structural Molecular Biology in the School of Molecular and Cellular Biology. Since 2014, his work has focused on how hydrodynamic forces can damage proteins, specifically, monoclonal antibodies. He studied biological chemistry at the University of Sheffield and obtained his Ph.D. at Leeds.
 Sheena Radford, Ph.D., OBE, FRS, FMedSci, is a biophysics professor at the University of Leeds’ Astbury Centre for Structural Molecular Biology in the School of Molecular and Cellular Biology. Her research focuses on protein folding and misfolding, amyloid, membrane protein folding, aggregation, and biopharmaceuticals. She has published more than 330 peer-reviewed papers and book chapters. She obtained her B.Sc. in biochemistry at the University of Birmingham and her Ph.D. at the University of Cambridge.
Sheena Radford, Ph.D., OBE, FRS, FMedSci, is a biophysics professor at the University of Leeds’ Astbury Centre for Structural Molecular Biology in the School of Molecular and Cellular Biology. Her research focuses on protein folding and misfolding, amyloid, membrane protein folding, aggregation, and biopharmaceuticals. She has published more than 330 peer-reviewed papers and book chapters. She obtained her B.Sc. in biochemistry at the University of Birmingham and her Ph.D. at the University of Cambridge.
 Nikil Kapur, Ph.D., is a professor of applied fluid mechanics at the University of Leeds in the School of Mechanical Engineering. He teaches students and industry about thermodynamics and fluid flow. He earned his B.Eng. at the University of Bradford and his Ph.D. at Leeds. He leads a group of researchers on a broad range of interdisciplinary projects spanning engineering, biology, and medicine and works closely with industry.
Nikil Kapur, Ph.D., is a professor of applied fluid mechanics at the University of Leeds in the School of Mechanical Engineering. He teaches students and industry about thermodynamics and fluid flow. He earned his B.Eng. at the University of Bradford and his Ph.D. at Leeds. He leads a group of researchers on a broad range of interdisciplinary projects spanning engineering, biology, and medicine and works closely with industry.
 David Brockwell, Ph.D., is a biochemistry and molecular biology professor at the University of Leeds’ Astbury Centre for Structural Molecular Biology in the School of Molecular and Cellular Biology. His research explores the effects that force has on proteins and membrane protein folding and folding factors with a focus on biopharmaceutical manufacturing. He obtained his B.Sc. and his Ph.D. at the University of Manchester.
David Brockwell, Ph.D., is a biochemistry and molecular biology professor at the University of Leeds’ Astbury Centre for Structural Molecular Biology in the School of Molecular and Cellular Biology. His research explores the effects that force has on proteins and membrane protein folding and folding factors with a focus on biopharmaceutical manufacturing. He obtained his B.Sc. and his Ph.D. at the University of Manchester.
