NIST, IBBR Researchers Explore Excipient-Influenced mAb Dynamics At Atomic Level
By Anupreet Kaur, Frank Delaglio, Subrata H. Mishra, John P. Marino, and Robert G. Brinson IBBR, NIST and the University of Maryland

Monoclonal antibodies (mAbs) are currently the leading platform for development of protein-based biological drugs due to their high specificity, binding properties, and developability. mAbs are used widely in the treatment of many illnesses, including inflammatory diseases, viral infections, autoimmune disorders, and cancer.
To ensure that the safety and efficacy of these drugs are maintained, clinically significant physiochemical parameters, known as critical quality attributes (CQA), are measured and monitored throughout the drug life cycle.
One CQA, higher order structure (HOS), is directly coupled to the function of protein biologics, and deviations in this CQA may be linked not only to a loss in effectiveness but can also result in adverse clinical events. These deviations may occur when certain solvent components affect higher rates of excursions to unfolded conformations, over time leading to undesirable quaternary structures, such as aggregation.
To maintain the therapeutic and safety profile of the drug substance, intensive formulation screening is performed during development using a battery of excipients (e.g., sugars, salts, amino acid, and surfactants). Such an approach often relies heavily on trial-and-error experimentation, making the process time-consuming and expensive.
Rational formulation development needs an understanding of how excipients influence biomolecular motion, which can then be leveraged to select conditions that enhance productive states that improve drug product stability. Additionally, knowledge of mAb motional trends at faster time scales (ps-ns, µs-ms), both in the presence and absence of formulation components, provides an opportunity to capture dynamic behavior that may result in long-term instability. To date, there have been no in-depth investigations of excipient-induced mAb dynamics at atomic-level resolution to better inform formulation selection. Here, we highlight how nuclear magnetic resonance spectroscopy (NMR) is a unique and powerful technique to investigate the structure and dynamics of the drug substance (DS) in various drug formulations. In-depth NMR investigations are now possible because of recent advances in expression of stable isotopically labeled mAbs. We illustrate how the understanding of DS-excipient interactions may ultimately provide a basis for rational formulation development.
Current Methods Offer Limited Resolution
Protein therapeutics are highly susceptible to physiochemical degradation pathways. Formulation serves the general purpose of promoting conformational, colloidal, and chemical stability by suppressing aggregation, reducing viscosity, enhancing chemical stability, and improving activity and bioavailability.1 Formulation scientists meticulously design formulations to maintain the integrity of mAbs throughout their shelf life and administration to patients.
To ensure the selection of the optimal formulation, accelerated stability studies are commonly performed to inform long-term physiochemical stability. A variety of high-throughput biophysical methods are used to evaluate potential formulations, including circular dichroism, differential scanning calorimetry, dynamic light scattering, Fourier transform infrared spectroscopy, and Raman spectroscopy. However, these techniques only afford low to moderate structural resolution, limiting the mechanistic understanding of how excipients influence mAb dynamics at the atomic level and how such dynamics correlate to macroscopic observations such as aggregation or stability.
To date, both one-dimensional (1D) and two-dimensional (2D) NMR methods at natural abundance have been developed extensively for atomic resolution fingerprinting of the biologic DS and drug product (DP).2 These methods have been extended to investigate the impact of excipients on DS, including arginine glutamate solutions,3 sugars,4 and polysorbates.5,6 Indeed, these methods have successfully related NMR parameters to overall conformational and colloidal stability, and 1D methods have been used to guide formulation screening.2
Despite these advances, the atomic-level information accessible by NMR has not been fully exploited due to the lack of assignments of the 1H, 13C, and 15N resonances of the protein DS. Indeed, while the 2D NMR fingerprinting method is seeing increased use during clinical development, without specific assignments any observed spectral perturbations can at best only be associated with residue type. Before 2023, only Fc backbone resonance assignments were known.7,8
After years of development, in 2023 the NMR group at the National Institute of Standards and Technology (NIST) reported the first-ever backbone assignments of a Fab domain, derived from the NISTmAb reference material 8671.9 Additional Fab domains were assigned by other laboratories in 2024.10,11 The NISTmAb, an IgG1κ, was developed for the pharmaceutical industry for purposes of assuring system suitability, benchmarking new analytical approaches, and harmonizing measurements.12 With resonance assignments in hand, the door has opened to in-depth NMR characterization of mAb-excipient interactions.
NMR Experiments Open Door To Probing mAb protein motions at an atomic scale
A more comprehensive understanding of the molecular structure and conformational flexibility of mAbs is needed to address the challenges associated with formulation screening. Missing from current formulation science is an in-depth biophysical understanding of excipient-induced protein dynamics at atom-level resolution. More in-depth knowledge of the interplay between mAb conformation and formulation would lead to knowledge-driven formulation designs and could reduce the overall time in the formulation development process.
With NMR resonance assignments in hand, specific biophysical properties of mAbs can be probed, including HOS (e.g., secondary, tertiary, and quaternary structure), conformational fluctuations, internal dynamics, and excipient-induced motions. Tertiary structure describes the spatial arrangement of the atoms within a protein, including the folding pattern of polypeptide chains and the positioning of the side chains. Quaternary structure defines the interaction regions of the different subunits of the antibody and intermolecular interactions, including aggregation. The conformational dynamics captures all the possible conformations of the antibody occurring in the solution. Interactions with excipients (whether specific or non-specific) or an antigen can alter mAb structure, dynamics, or both. Changes in local motions greatly impact overall protein dynamics and propensity to aggregate. For instance, at higher temperatures, alterations in protein local motions might expose previously buried hydrophobic patches to the solvent. Aggregation is more likely when this exposure happens on a timescale similar to the rate of molecular collisions, and it is enhanced at higher protein concentrations. Altering local motions also alters molecular interactions and binding, in much the same way as a single amino acid mutation in hemoglobin causes the aggregation that leads to sickle cell anemia.13
Not surprisingly, temperature, higher concentrations, and mutations affect protein stability, and successful formulations should alter deleterious motions that increase aggregation propensity. However, to rationally employ these ideas we have to detect local atomic motions of the protein backbone in a wide variety of time scales and correlate them to global domain motions.
Motions at different time scales are manifested in several different NMR measurables. The standard set of NMR dynamics experiments measures the longitudinal relaxation rate (R1), transverse relaxation rate (R2), and heteronuclear steady-state NOE of the backbone amide nitrogen. This suite of experimental measurements reports on protein backbone dynamics at the picosecond to nanosecond timescale. R1ρ spin-lock and Carr-Purcell-Meiboom-Gill (CPMG) spin-echo NMR experiments are commonly used for probing the conformational dynamics of macromolecules at the microsecond to millisecond timescale. NMR experiments can also probe global parameters such as size, shape, and aggregation status that can be obtained from translational diffusion coefficients and rotational correlation times derived from diffusion-ordered spectroscopy (DOSY) and transverse relaxation optimized spectroscopy for rotational correlation times (TRACT) measurements respectively.
While there is no precedent for studying mAb therapeutics with these methods, they have been successfully applied to the small therapeutic protein filgrastim in various formulations14 and are commonly applied to other proteins of academic and clinical interest.15,16
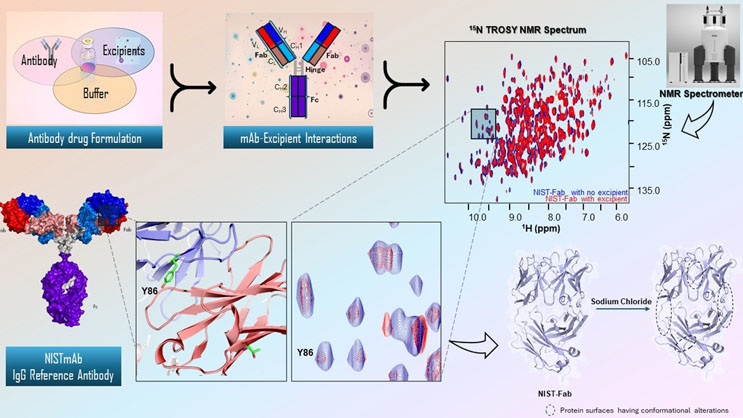
The NIST-UMD NMR group is inaugurating in-depth biophysical NMR characterization of therapeutic mAb-excipient interactions and dynamics. Specifically, using the amide backbone signal assignments of the NIST-Fab, which allow us to track each backbone amide 15N atom, we are focusing on 15N relaxation and diffusion experiments for studying NIST-Fab in the presence of various pharmaceutically relevant formulations. These in-depth NMR analyses with common excipients – sodium chloride, sucrose, and polysorbate 80 – have revealed how each excipient alters local (atomic) and global (shape) Fab dynamics (Figure 1, bottom right). With these initial experiments completed, efforts can turn to understanding the underlying mechanisms of aggregation induced, or the stability conferred, by specific excipients.
Figure: The schematic shows how NMR spectroscopy provides atomic-level resolution to characterize mAb structure and dynamics. It emphasizes the technique's ability to reveal detailed mAb dynamics in the presence of excipients. Click for full size.
Challenges of Atomic Scale NMR Include Requirements and Costs of Expression System for Isotopic Labeling
Monoclonal antibodies are large macromolecules (around 150 kDa) with a large number of peaks in the spectrum. Before any investigation into molecular motions can occur, each of these peaks must be first assigned to its corresponding atom. The assignment process necessitates higher-dimensional NMR experiments, which cannot be performed on mAb samples at natural isotopic abundance. Further, to simplify the assignment process, often large multi-domain proteins are split into their constituent domains and have their backbone signals assigned separately. However, for a therapeutic protein, the individual domains must maintain the integrity of HOS compared to the intact mAb. For the NISTmAb, this HOS integrity was demonstrated by the direct overlay of signals from the constituent domains onto a reference spectrum on the intact mAb.17
Another requirement for in-depth NMR structural studies is mAbs with 2H, 13C, and 15N isotopic labeling. A major hindrance in the advancement of this field has been the lack of expression systems for production of these isotopically labeled materials. However, after years of development, as of 2024 there are now multiple expression systems published, including the yeast k. phaffii,18 e. coli,19 and a cell-free system.11 In addition to the NIST-Fab, resonance assignments are now available for other Fab domains, including adalimumab,10 trastuzumab,20 and a therapeutic Fab against the human LAMP1 glycoprotein.11 Once assignments have been made, ideally under pharmaceutically relevant conditions, these can be directly transferred to spectra of therapeutic mAbs recorded at natural isotopic abundance.
While production of deuterated Fab domains has been solved, to date intact mAbs have only been labeled with 13C and 15N at great expense, such as by adding isotope labeled amino acids into the media during CHO expression. Although signal assignments can be transferred from the constituent domains to the intact mAb, development of less expensive expression systems continues for intact mAbs, with the additional issue that the glycosylation pattern generated by the expression systems will likely be different from the mAb drug substance. As the glycosylation pattern is a CQA, this issue will need to be addressed once a suitable expression system has been developed. However, preliminary evidence suggests that glycan patterns primarily affect mAb dynamics, that is, the chemical shifts of relevant resonances near the glycans change very little, if at all; it is the peak linewidth that primarily changes.21,22 As such, it may be possible that chemical shifts may be confidently mapped from the isotopically labeled mAb to the therapeutic mAb, even if the glycan distributions are different.
Conclusion
Although the application of NMR to formulation science is in its infancy, the atom-level detail afforded by these in-depth methods promises to be transformative. NMR is indeed at a watershed moment: in the past year, three different laboratories have developed three different workable expression systems. Two of the systems afforded 13C methyl specific labeling and methyl group assignments. As the 2D NMR fingerprinting method relies on the methyl fingerprint, transferring the findings from the constituent domains to the intact mAbs should become practical.
Furthermore, once a library of mAb-excipient results has been established, implementing machine learning predictive algorithms, in combination with other synergistic analytical data, will become possible.
Ultimately, a more in-depth knowledge-based approach to formulation design driven by advanced NMR measurements could significantly reduce the time and expense associated with developing stable, effective mAb therapies.
Disclaimer: Certain commercial equipment, instruments, and materials are identified in this paper in order to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the material or equipment identified is necessarily the best available for the purpose.
References:
- Razinkov, V. I.; Treuheit, M. J.; Becker, G. W., Accelerated formulation development of monoclonal antibodies (mAbs) and mAb-based modalities: review of methods and tools. J Biomol Screen 2015, 20 (4), 468-83.
- Ma, J.; Pathirana, C.; Liu, D. Q.; Miller, S. A., NMR spectroscopy as a characterization tool enabling biologics formulation development. Journal of pharmaceutical and biomedical analysis 2023, 223, 115110.
- Kheddo, P.; Cliff, M. J.; Uddin, S.; van der Walle, C. F.; Golovanov, A. P., Characterizing monoclonal antibody formulations in arginine glutamate solutions using (1)H NMR spectroscopy. MAbs 2016, 8 (7), 1245-1258.
- Zhang, C.; Gossert, S. T.; Williams, J.; Little, M.; Barros, M.; Dear, B.; Falk, B.; Kanthe, A. D.; Garmise, R.; Mueller, L.; Ilott, A.; Abraham, A., Ranking mAb-excipient interactions in biologics formulations by NMR spectroscopy and computational approaches. MAbs 2023, 15 (1), 2212416.
- Poppe, L.; Knutson, N.; Cao, S.; Wikstrom, M., In Situ Quantification of Polysorbate in Pharmaceutical Samples of Therapeutic Proteins by Hydrodynamic Profiling by NMR Spectroscopy. Anal Chem 2019, 91 (12), 7807-7811.
- Singh, S. M.; Bandi, S.; Jones, D. N. M.; Mallela, K. M. G., Effect of Polysorbate 20 and Polysorbate 80 on the Higher-Order Structure of a Monoclonal Antibody and Its Fab and Fc Fragments Probed Using 2D Nuclear Magnetic Resonance Spectroscopy. Journal of pharmaceutical sciences 2017, 106 (12), 3486-3498.
- Yagi, H.; Zhang, Y.; Yagi-Utsumi, M.; Yamaguchi, T.; Iida, S.; Yamaguchi, Y.; Kato, K., Backbone (1)H, (13)C, and (15)N resonance assignments of the Fc fragment of human immunoglobulin G glycoprotein. Biomol NMR Assign 2015, 9 (2), 257-60.
- Liu, D.; Ren, D.; Huang, H.; Dankberg, J.; Rosenfeld, R.; Cocco, M. J.; Li, L.; Brems, D. N.; Remmele, R. L., Jr., Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry 2008, 47 (18), 5088-100.
- Solomon, T. L.; Chao, K.; Gingras, G.; Aubin, Y.; O'Dell, W. B.; Marino, J. P.; Brinson, R. G., Backbone NMR assignment of the yeast expressed Fab fragment of the NISTmAb reference antibody. Biomol NMR Assign 2023, 17 (1), 75-81.
- Sarker, M.; Aubin, Y., Backbone and methyl side-chain resonance assignments of the Fab fragment of adalimumab. Biomol NMR Assign 2024.
- Giraud, A.; Imbert, L.; Favier, A.; Henot, F.; Duffieux, F.; Samson, C.; Frances, O.; Crublet, E.; Boisbouvier, J., Enabling site-specific NMR investigations of therapeutic Fab using a cell-free based isotopic labeling approach: application to anti-LAMP1 Fab. Journal of biomolecular NMR 2024, 78 (2), 73-86.
- Schiel, J. E.; Turner, A.; Mouchahoir, T.; Yandrofski, K.; Telikepalli, S.; King, J.; DeRose, P.; Ripple, D.; Phinney, K., The NISTmAb Reference Material 8671 value assignment, homogeneity, and stability. Anal Bioanal Chem 2018, 410 (8), 2127-2139.
- Gabriel, A.; Przybylski, J., Sickle-cell anemia: A Look at Global Haplotype Distribution. Nature Education 2010, 3 (3), 2.
- Ghasriani, H.; Frahm, G. E.; Johnston, M. J. W.; Aubin, Y., Effects of Excipients on the Structure and Dynamics of Filgrastim Monitored by Thermal Unfolding Studies by CD and NMR Spectroscopy. ACS Omega 2020, 5 (49), 31845-31857.
- Sekhar, A.; Kay, L. E., An NMR View of Protein Dynamics in Health and Disease. Annu Rev Biophys 2019, 48, 297-319.
- Stenstrom, O.; Champion, C.; Lehner, M.; Bouvignies, G.; Riniker, S.; Ferrage, F., How does it really move? Recent progress in the investigation of protein nanosecond dynamics by NMR and simulation. Current opinion in structural biology 2022, 77, 102459.
- Arbogast, L. W.; Brinson, R. G.; Marino, J. P., Mapping monoclonal antibody structure by 2D 13C NMR at natural abundance. Anal Chem 2015, 87 (7), 3556-61.
- Chao, K. L.; O'Dell, W. B.; Solomon, T. L.; Brinson, R. G.; Marino, J. P.; Kelman, Z., Expression of 2H, 13C, 15N-labeled NIST-Fab Fragment in the Methylotrophic Yeast Komagataella Phaffii for Nuclear Magnetic Resonance Studies. EPJ Web Conf. 2023, 286, 01003.
- Gagne, D.; Sarker, M.; Gingras, G.; Hodgson, D. J.; Frahm, G.; Creskey, M.; Lorbetskie, B.; Bigelow, S.; Wang, J.; Zhang, X.; Johnston, M. J. W.; Lu, H.; Aubin, Y., Strategies for the production of isotopically labelled Fab fragments of therapeutic antibodies in Komagataella phaffii (Pichia pastoris) and Escherichia coli for NMR studies. PLoS One 2023, 18 (11), e0294406.
- Gagne, D.; Aramini, J. M.; Aubin, Y., Backbone and methyl side-chain resonance assignments of the single chain Fab fragment of trastuzumab. Biomol NMR Assign 2024.
- Arbogast, L. W.; Delaglio, F.; Schiel, J. E.; Marino, J. P., Multivariate Analysis of Two-Dimensional 1 H, 13 C Methyl NMR Spectra of Monoclonal Antibody Therapeutics To Facilitate Assessment of Higher Order Structure. Anal Chem 2017, 89 (21), 11839-11845.
- Barb, A. W.; Prestegard, J. H., NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat Chem Biol 2011, 7 (3), 147-53.
About The Authors:
 Anupreet Kaur is post-doctoral research fellow at NIST-IBBR, focusing on elucidating the biophysical mechanisms of pharmaceutically relevant formulations and exploring the dynamics of monoclonal antibodies at atomic level using advanced NMR. She earned her Ph.D. from Guru Nanak Dev University, India and then joined George Washington University where she performed protein engineering to enhance ligase activity. She can be contacted at akaur129@umd.edu.
Anupreet Kaur is post-doctoral research fellow at NIST-IBBR, focusing on elucidating the biophysical mechanisms of pharmaceutically relevant formulations and exploring the dynamics of monoclonal antibodies at atomic level using advanced NMR. She earned her Ph.D. from Guru Nanak Dev University, India and then joined George Washington University where she performed protein engineering to enhance ligase activity. She can be contacted at akaur129@umd.edu.
 Subrata H. Mishra is a principal investigator in the Biomolecular Structure and Function Group under the Biomolecular Measurement Division at NIST. He is stationed at the IBBR. His research at NIST/IBBR aims to translate our understanding of biomolecular dynamics into real-world applications ranging from biosensors to biotherapeutics. Additionally, his research interest encompasses NMR method development and metabolomics measurements in biomanufacturing. He earned his B.E. in chemical engineering from the University of Mumbai and his Ph.D. in chemistry from Georgia State University. He can be reached at subrata.mishra@nist.gov.
Subrata H. Mishra is a principal investigator in the Biomolecular Structure and Function Group under the Biomolecular Measurement Division at NIST. He is stationed at the IBBR. His research at NIST/IBBR aims to translate our understanding of biomolecular dynamics into real-world applications ranging from biosensors to biotherapeutics. Additionally, his research interest encompasses NMR method development and metabolomics measurements in biomanufacturing. He earned his B.E. in chemical engineering from the University of Mumbai and his Ph.D. in chemistry from Georgia State University. He can be reached at subrata.mishra@nist.gov.
 Frank Delaglio is a principal investigator at the National Institute of Standards and Technology, developing computational methods for NMR data to support development and manufacturing of drugs and vaccines and to support basic research in structural biology. Frank's career includes commercial software development, extensive pharma consulting, and regular participation as an advanced course instructor for the European Molecular Biology Organization. Frank earned a B.A. in chemistry from Syracuse University, and a Ph.D. from the Osaka University Graduate Department of Pharmaceutical Sciences. He can be reached at frank.delaglio@nist.gov.
Frank Delaglio is a principal investigator at the National Institute of Standards and Technology, developing computational methods for NMR data to support development and manufacturing of drugs and vaccines and to support basic research in structural biology. Frank's career includes commercial software development, extensive pharma consulting, and regular participation as an advanced course instructor for the European Molecular Biology Organization. Frank earned a B.A. in chemistry from Syracuse University, and a Ph.D. from the Osaka University Graduate Department of Pharmaceutical Sciences. He can be reached at frank.delaglio@nist.gov.
 John P. Marino is co-director of the IBBR, a joint research institute of the University of Maryland and NIST and the group leader of the NIST Biomolecular Structure and Function Group at IBBR. John’s research focuses on the application of NMR and other biophysical methods to advance measurement of proteins and nucleic acids, with a particular focus on application to biotherapeutics and vaccines. John received an A.B. in chemistry from Princeton University and a Ph.D. in chemistry from Yale University. He can be contacted at john.marino@nist.gov.
John P. Marino is co-director of the IBBR, a joint research institute of the University of Maryland and NIST and the group leader of the NIST Biomolecular Structure and Function Group at IBBR. John’s research focuses on the application of NMR and other biophysical methods to advance measurement of proteins and nucleic acids, with a particular focus on application to biotherapeutics and vaccines. John received an A.B. in chemistry from Princeton University and a Ph.D. in chemistry from Yale University. He can be contacted at john.marino@nist.gov.
 Robert G. Brinson is currently a principal investigator in the Biomolecular Structure and Function Group at NIST in Rockville, Maryland. Robert applies biophysical measurements to define structure and dynamics of nucleic acids, proteins, and glycoproteins. In particular, Robert seeks to push the limits of NMR methodologies to define the critical quality attribute of higher order structure for biologics. Robert received B.A. in chemistry and music from Houghton College and his Ph.D. in chemistry from Wake Forest University. He can be reached at robert.brinson@nist.gov.
Robert G. Brinson is currently a principal investigator in the Biomolecular Structure and Function Group at NIST in Rockville, Maryland. Robert applies biophysical measurements to define structure and dynamics of nucleic acids, proteins, and glycoproteins. In particular, Robert seeks to push the limits of NMR methodologies to define the critical quality attribute of higher order structure for biologics. Robert received B.A. in chemistry and music from Houghton College and his Ph.D. in chemistry from Wake Forest University. He can be reached at robert.brinson@nist.gov.